Massbalance computing (Stocks, fluxes, budgets)¶
- This functionnality allows you to compute for each substance :
- fluxes through boundaries
- budgets (stocks and fluxes ) in sub-domains
- By default, one domain is considered, containing all the computational grid. Or user can define one or several subdomain and boundaries in a specific file, with a name different from “none”.
- To use this functionnality : set l_bilan=.true. in parasubs.txt : namelist for substances , associated with the number of border (open and closed), the output time step, the start date of massbalance computing (since version r2097), the file name where zones and borders are defined if needed. (parasubs.txt : namelist for substances )
What it calculates ?¶
- For closed sub-domain (pink area on figure):
- Net cumulated fluxes (since the start date of massbalance computation) in and out of a given sub-domain
- Flxcum=∑t(FlxW−FlxE+FlxS−FlxN)
- Net cumulated input fluxes into the sub-domain due to rivers discharges and exchanges with atmosphere.
- Flincum=∑t(Flxrej+Flxatm)
- If key_contaminant : cumulated fluxes by exchange between substances due to biogeochemical reactions
- Stocks in the water column and in the sediment within the sub-domain
- For open boundaries (green line on figure):
- Net cumulated fluxes through boundaries
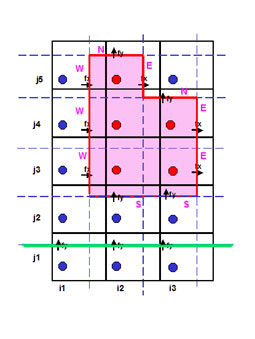
- boundaries and sub-domains are defined from the surface water to the bottom
- a budget sub-domain is only valid if the boundary is closed (massbalance computing)
- Budget : Bud=Stockwater+Stocksed−Flxcum−Flincum : must be constant if conservative substance
How to buid the file which defines sub-domain ?¶
one example of file defining sub-domains and boundaries : subbudgetdomain.dat is available in $HOMEMARS/../EXAMPLES/
the name of this file is set in parasubs.txt : namelist for substances (name_bilfil)
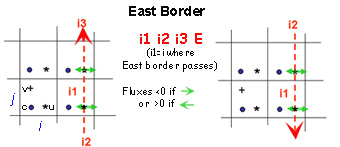
Boundaries are defined by several contiguous segments S-N and/or W-E (any number of segments and any direction (S-N or N-S, WE or E-W).
- Each segment is characterized by 3 indices and a letter N, S, W or E to determine whether this border is a north, south, west or east segment of the sub-domain.
- For a closed sub-domain : “East” segments are borders placed at the east of the closed area. “South” segments are borders placed at the south of the area etc..
- For an open boundary, E,W,S,N give the direction of flow
For north and south segments, the 3 indices are i1=i_start ; i2=i_end and i3=j
For west and east segments, the 3 indices are i1=i ; i2=j_start ; i3=j_end
Start and end indices of meshes that define segments correspond to the meshes in which there are the borders (see figures below)
Warning
These rules are valid for newer versions (>Version r2097 - decembre2015).Indeed, in older versions, the start and end indices of meshes that define South and West segments correspond to meshrespectively located right above (j+1) or just east (i+1) the mesh in which crosses the border.
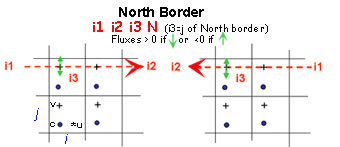
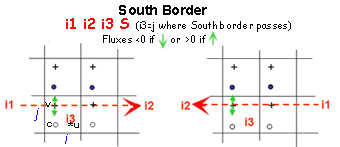
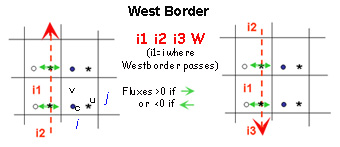
Budgets are computed only inside sub-domains surrounded by closed boundaries
ASSIGN open boundaries FIRST and closed boundaries next
Note
Verification of domains¶
- You can verify your closed domains in the output file fic_verif_zone_budget created at the beginning of the simulation.
- A script Python, ficverif_zone_budget.py created by B. Mengual, is proposed in $HOMEMARS/../TOOLS/PARSERVAR/ in order to read this file and see the area directly in a figure and in your bathymetry.
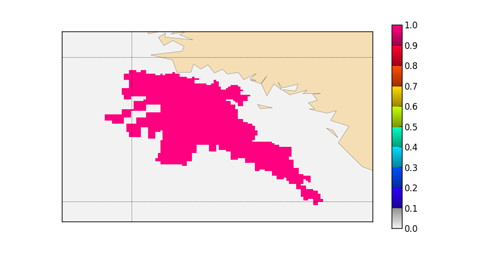
- Example of mask_bud in a closed aera :
- Example of mask for a complex budget area, drawn by the script python from B. Mengual (Grande Vasiere : mask_bud=1 ==> pink color)
Massbalance output Files¶
- results are written in output files .csv (separator = ‘;’)
- 1 file for each budget sub-domain (closed boundary) and 1 file each 10 variables (1 file for the first 10 variables; second file for variables 10 to 20...)
- 1 file for each open boundary (fluxes through each boundary)
- into the file : One line /time_out_budget
- Massbalance_output_filenames are automatically created :
- budget sub-domain : outputfilename_suffix_bil[boundary number]_1.csv (1 for the first file)
- fluxes through open boundary : outputfilename_suffix_border[boundary number].csv
- inputs of substances are observed if the obc cumulated fluxes increase as a function of time (positive slope) ; and vice versa.
Tracking points¶
This functionnality is useful to save vertical profiles in only a few points and with a shorter time step than in output file
- Define the following variables inside paraspec.txt :
- l_points=.true.
- file_point=’../inputs/points.dat’
- Define tracking points in the file : points.dat (under inputs)
points.dat :
+----------------------------------+----------------------------------------------------------------+ | ******************************* | **************************************************** | | Point1 | prefix of output file name | | 06/03/1900 00:00:00 | starting date | | 06/03/2500 00:00:00 | ending date | | 600.0 | time step (s) | | .true. | location in i,j,k (.true.) or in lon,lat,k (.false.) | | 29 29 12 | location | | .true. | l_out_ssh_track | | .false. | l_out_ubt_track | | .true. | l_out_u3d_track | | .true. | l_out_sal_track | | .true. | l_out_temp_track | | ******************************* | **************************************************** | +----------------------------------+----------------------------------------------------------------+
Diagnostic variables : vardiag.dat¶
- Diagnostic variables are chosen and computed by the user in the subroutine : diagnostic.F90 or elsewhere, in order to save some diagnostics or indicators
- if l_diag =.true. in paraspec.txt and key_biolo
- these variables are not used during simulation but are written in a specific output file in addition to the file with concentrations of the state variables.
- Definition and names of diagnostic variables are set in a file vardiag.dat. The name of this file vardiag.dat (file_diag) is set in paraspec.txt
- start time and end time for calculate diagnostic variables are set in paraspec.txt
Warning : this functionnality (with writing in the diagnostic output file) is available only with the key key_biolo
- vardiag.dat (model in $HOMEMARS/../EXAMPLES/vardiag.dat) :
+----------------------------------------------------+----------------------------------------------------------------+ | ************************************************ | **************************************************** | | 1 | number of the diagnostic variable | | grad_vert_salinite | variable name | | grad_vert_salinite | variable long_name | | maximum_vertical_gradient_of_sea_water_salinity | variable standard_name | | per mil | unit | | 0.0 | variable valid_min value | | 100.0 | variable valid_max value | | 2 | dimension (1=1D ; 2=2D ; 3=3D) | | .true. | variable in water (k,i,j) | | .false. | variable in sediment (i,j,k) | | .true. | saving in file | | *********************************************** | **************************************************** | +----------------------------------------------------+----------------------------------------------------------------+
Driving and Intermediate variables¶
Driving variables are computed by the user in the subroutines : user_init_vdriv and user_ftime_vdriv in the module usersubstance.F90
their names and definition are set in variable.dat : Defined simulated substances
they are initialized by the user in user_init_vdriv.
- The user must set this initialization in order to give values at each point of the area, | by programming himself a function and if needed reading a file
If l_driv_cst =.false., driving variables are not constant.
They are computed in the subroutine user_ftime_vdriv ; interpolated (every dt_driv time step) or computed again or read again from an input file , progammed by the user.
- Intermediate variables are computed by the user in the subroutines : user_vintermediate in the module usersubstance.F90
- their names and definition are set in variable.dat : Defined simulated substances
- they are useful when you need to calculate at each point a combination of variables and parameters.
- The function has to be programmed by the user in user_vintermediate
